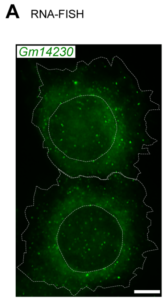
Current Major Research Themes Juvenile Biology Stem Cell–Based Regenerative Medicine Rapid Whole-Genome Diagnosis Neurodevelopment and Hypothalamic Maturation Parent–Offspring Bond Formation Cell Competition Therapeutic Studies of Pediatric Intractable Diseases Juvenility Different from adults, children make tremendous changes such as growth, maturation, adaptation, regeneration, and so on. How are they equipped for children? Are they equipped in children's cells? Children suffering from diseases occasionally show a slowdown of growth (growth retardation) but also 'catch-up growth' after eradicating diseases. The catch-up growth can be even faster than the normal growth speed. That is, children harbor a hidden potential for growth that can be induced if the conditions are met. This is where the ideas of inducing the juvenile property for therapeutic purposes come from. Cellular Juvenescence Cellular juvenescence is a feature of cells residing in juvenile animals (including humans). The juvenile cells tend to proliferate rigorously, maintain the potential for functional maturation and differentiation, grow in size, and show resistance against undergoing senescence. In cultured cells, we identified Gm14230, SRSF7, and BEX1 were essential to maintain cellular senescence. When the functions of these genes are suppressed, cells can no more maintain the cellular juvenescence, stop growth, lose differentiation capacity, and show hallmarks of senescent cells. JAGs Juvenility-associated genes (JAGs) are the group of genes that are selectively highly expressed during juvenile. Though initially identified in mice, they are well conserved in humans. What are the functions of the JAGs? How do they realize the juvenile properties in cells and animals? Among the JAGs, we have identified the functions of SRSF7 in age-dependent alternative splicing (ADAS). We'd like to explain ADAS in the section later. We also found the role of BEX1 in making the primary cilia. We discovered the fundamental role of BEX1 in tubulin polymerization and identified BEX1 was indispensable cilia formation. Cilia are antenna-like cellular structures through which cells can sense the external world. The concentration of nutrients (or any ligands), the flow of extracellular fluids, and stiffness of the extracellular matrix (ECM), all function as signaling inputs to cells via cilia. As we demonstrated with Bex1 knockout mice, cilia defects lead to a compromised sensing mechanosensing by renal tubular cells, ending in the dilation of renal tubules (polycystic disease of the kidney). For the physical aspect of BEX1 protein, we explain it in the "phase separation" section. BEX1 paper is freely available from here JALNCs Long noncoding RNAs (lncRNAs) are the RNA species that do not harbor a potentially protein-coding sequence longer than 200 nucleotides. Their mechanisms of action have been elusive in a large part. JALNCs are the lncRNAs that are selectively highly expressed in juvenile cells. Among the comprehensively identified JALNCs, we discovered that JALNC "Gm14230" was highly evolutionarily conserved and functioned to maintain cellular juvenescence. The subcellular localization of Gm14230 lncRNA is both in the cytosol and the nucleus as shown by RNA-fluorescent in situ hybridization (FISH) (Picture "RNA-FISH"). Please find the JALNC paper here ADAS We for the first time demonstrated the alternative splicing (AS) underwent in the postnatal growth phase. Using a mouse model, we uncovered that juvenile cells operate AS so that they can produce "anabolic" variants of genes. For example, microRNA-dependent translation suppressor EIF4A2 is expressed as the form of the "anabolic" type, rather than the "catabolic" type that is expressed in adults. We comprehensively identified the juvenile-specific AS as "age-dependent alternative splicing" (ADAS). ADAS paper is freely accessible from here Cell Competition Cell competition is a system that multicellular organisms operate to eradicate environmentally unfit cells. Evolutionarily conserved from flies, cell competition functions to remove precancerous cells and cells undergoing maldevelopment. How are the "unfit" cells detected by the system? It is now being revealed the presence of surrounding cells and sensing surface molecules and mechano-stimuli are crucial. Cytoophidia We discovered that TBC1D24, a protein associated with epileptic encephalopathy, forms a cellular macrostructure called cytoophidia. The TBC1D24 cytoophidia take ring or rod form (Picture). The formation of cytoophidia was provoked by the loss of cellular juvenescence, suggesting the cytoophidia formation was counteracted in juvenile cells. Phase separation
Phase separation separates the cellular interiors into compartments. We revealed that BEX1 protein harbors the physicochemical property as an intrinsically disordered protein (IDP) and is sufficient to form biomolecular condensates that facilitate tubulin polymerization in the reconstituted system. BEX1 localizes to pericentriolar material (PCM) at the bases of primary cilia and is indispensable for ciliogenesis both in vitro and in vivo.
In children, distinct gene sets are expressed compared with adults, giving rise to organelle functions and cellular behaviors unique to early life. These juvenility-associated genes (JAG) act through diverse mechanisms—including age-dependent alternative splicing, the formation of juvenile compartments, and the linear mitochondrial segment (LMS)—to orchestrate growth, development, and regeneration that are absent in adulthood. Mutations in JAGs frequently underlie pediatric diseases. By elucidating juvenile molecular mechanisms, we aim to uncover the causes of currently intractable childhood disorders and pave the way for novel therapies.
iPS cells, ES cells, and tissue stem cells hold great promise for regeneration. During childhood, numerous tissue-resident or stem-cell-like populations drive organ growth, adaptation, and repair. By dissecting the molecular basis of cellular juvenescence, we focus on these juvenile stem-like cells to advance our understanding of pediatric cancers and to develop innovative regenerative technologies.
Leveraging recent advances in genome sequencing and informatics, we are establishing ultra-fast whole-genome analysis pipelines that accelerate diagnosis and illuminate molecular disease mechanisms. This endeavor integrates the expertise of clinicians and researchers across the National Center for Child Health and Development.
We investigate the molecular underpinnings of neurodevelopmental disorders using state-of-the-art techniques: single-cell sequencing, spatiotemporal transcriptomics, optogenetics, fiber photometry, and detailed behavioral assays. Our goal is to decode the maturation programs of the hypothalamus and broader neural circuits.
To survive in nature, young mammals possess unique physiological traits that facilitate bonding with caregivers. Using mouse models, we analyze diverse developmental milestones during the neurodevelopmental period to reveal the neural mechanisms that motivate parental caregiving behaviors.
Multicellular organisms maintain tissue integrity through cell competition. We focus on juvenile-specific competitive interactions, aiming to unravel how three-dimensional cell assemblies preserve overall homeostasis via mechanical and molecular pathways.
Genome-wide association studies identify numerous variants linked to disease, yet functional validation is often lacking. By rapidly creating genome-edited model organisms, we scrutinize candidate variants to pinpoint therapeutic targets and accelerate drug discovery for rare pediatric conditions.
We collectively call the children's inherent properties including growth potential "juvenility" and are trying to understand the molecular machinery underlying it. So far, we have identified the genes selectively expressed during juvenile. The genes as we call "juvenility-associated genes" or "JAGs" are considered to function as building blocks for juvenility in cells.
We also have identified several genes that are necessary to maintain juvenility in cells. Without the genes, the cells lose "cellular juvenescence".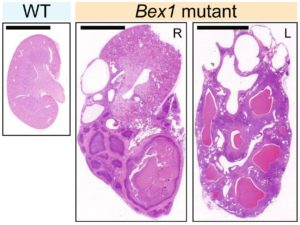
An age-dependent RNA editing modifies Gm14230. The Gm14230 RNA is a substrate for the Adenosine-Inosine (A to I) RNA editing, which is predominantly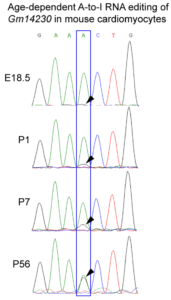 observed in older cells (Picture showing chromatogram of the RNA sequencing).
observed in older cells (Picture showing chromatogram of the RNA sequencing).
The function of Gm14230 is to suppress the transcription of TGIF2 via recruitment of EZH2, a histone methyltransferase.
Without Gm14230, cells undergo premature senescence, suggesting that Gm14230 functions to resist the occurrence of cellular senescence.
Furthermore, we identified SRSF7 as an executor of ADAS. The minigene assay demonstrated that deletion of the SRSF7 gene lead to switching from the juvenile to the adult type of AS. SRSF7-mutant mice exhibit fast-forwarded ADAS, ending in premature growth-exit and developmental failures. In the juvenility context, we found that the cellular juvenescence is compromised in the cells that are determined to take the loser cell fate. The loser cells in the in vitro setting modeling the neuroepithelium are induced to undergo p38-dependent apoptosis and eventually phagocytosed by surrounding cells. As the pivotal genes that mediate the juvenescence in the context above, we identified SRSF7 and EZH2.
In the juvenility context, we found that the cellular juvenescence is compromised in the cells that are determined to take the loser cell fate. The loser cells in the in vitro setting modeling the neuroepithelium are induced to undergo p38-dependent apoptosis and eventually phagocytosed by surrounding cells. As the pivotal genes that mediate the juvenescence in the context above, we identified SRSF7 and EZH2.
Please find the paper here.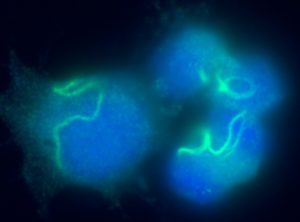
Please find the cytoophidia paper here.
For more detail, please find the paper here.
